Getting Started¶
ChemLG generates molecules based on the following combinatorial ‘linking’ and ‘fusion’ schemes:
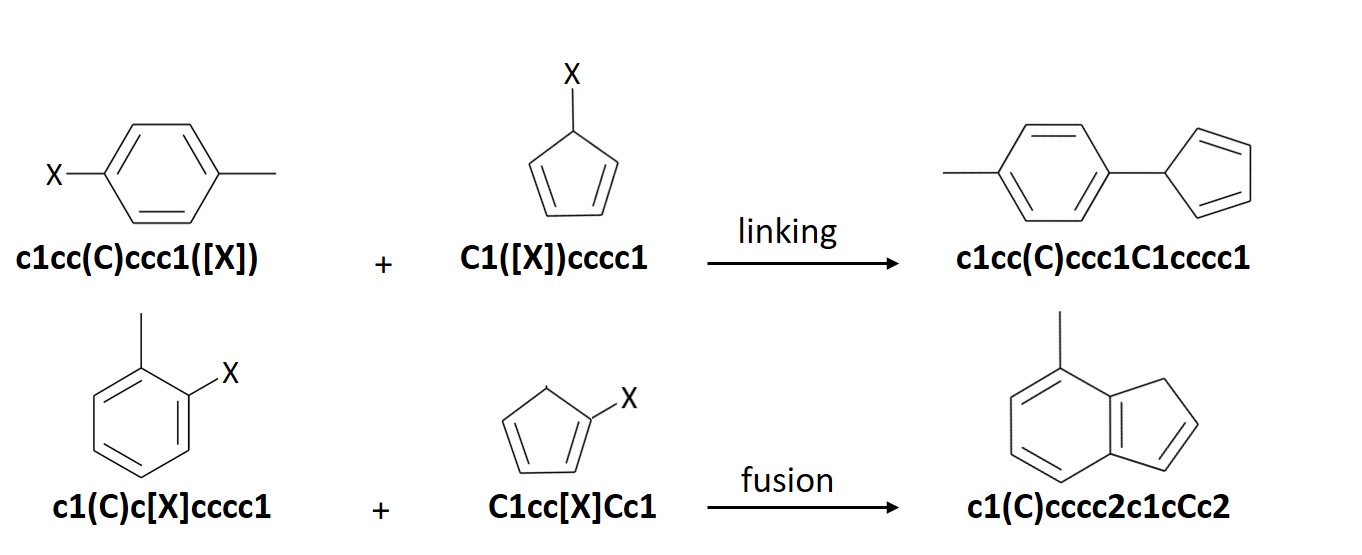
The ChemLG module is executed from the following command line:
chemlgshell -i ./config.dat -b ./building_blocks.dat -o ./output/
The file names used above are the default values that the program looks for. The -o handle is used to specify the output directory where all the output files will be dumped. As seen from the command line above, the user should provide two input files:
a file with the configuration/rules for generating the library with the -i handle
a file with all the initial building blocks with the -b handle
Templates for both the input files are located at https://github.com/hachmannlab/chemlg/tree/master/chemlg/templates
It is recommended that the user substitutes the required values directly into the respective templates for the input files in order to avoid any conflicts later while executing the program. For more specifics into how to make the input files, look at:
ChemLG Graphical Input File Builder¶
We also offer a way to build the input files via a graphical user interface through a jupyter notebook.
The following additional packages should be installed in the virtual environment before running the GUI builder. The last command below adds a “chemlg” kernel to the jupyter notebook.
conda install –c rdkit rdkit
conda install ipykernel
conda install -c conda-forge ipywidgets
python -m ipykernel install --name my_chemlg_env --display-name "chemlg"
To use the GUI, launch jupyter notebook and enter:
from chemlg.notebooks.main import config_builder
config_builder()
When the user is done building the input files, the user also has an option to run the code directly from the jupyter notebook.
Constrained library generation via Genetic Algorithm¶
ChemLG also offers the possibility for constrained growth of a library. That is, a library can be generated such that new members are added to the library only if they conform to a given range of target properties. For example, a library with each of the members having a density greater than 1000 kg/m3. The constrained library can also be made to optimize multiple properties at once using the multi-objective Genetic Algorithm module.
To generate this constrained library using Genetic Algorithm, the user has to specify the following:
desired range of target property/properties
an objective function
Genetic Algorithm parameters
For example:
Define the objective function:
# File name: example_obj.py
import pybel
# This 'objective' function always receives an openbabel object of a molecule ('mol_ob' in the example).
def objective(mol_ob):
mol2 = pybel.readstring("smi", "c1ccccc1")
tanimoto = mol_ob.calcfp()|mol2.calcfp()
# tanimoto is the desired property which is returned by the function
return tanimoto
Execute ChemLG via Genetic Algorithm:
# File name: constrained_lib.py
from chemlg.constrained import GeneticAlgorithm # import genetic algorithm
from example_obj import objective # import objective function
ga_library = GeneticAlgorithm(evaluate=objective,
fitness = (("max", 0.7), )
bb_file = <specify building block file handle here>,
config_file = <specify config file handle here>,
output_dir = <specify output directory here>)
best_ind_df, best_individual = ga_library.search(n_generations=20)
For more information on Genetic Algorithm parameters, refer to https://github.com/hachmannlab/chemlg/blob/master/chemlg/constrained.py
Feasibility Analysis¶
Even with the user-defined constraints that help eliminate undesired molecules, the final library can still end up with molecules that are not synthetically feasible. To address this issue of feasibility, we have incorporated a code that ranks these virtual molecules against a database of commercially available compounds (obtained from MolPort) based on the similarity of their fingerprints. With an upper or lower bound to the Tanimoto index provided by the user, the code processes the final library to remove all the molecules that fall short of the given criteria.
The following similarity metrics are included in ChemLG:
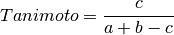
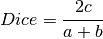
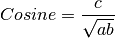
where, a is the number of features in structure 1,
b is the number of features in structure 2,
and c is the number of common features in structures 1 and 2.
Openpyxl is required as an additional dependency in ChemLG to run the feasibility analysis and can be installed as:
pip install openpyxl
The input to this function is the library generated by ChemLG and the cutoff Tanimoto index. This also gives the statistics of the generated library.
To run the feasibility analysis:
from chemlg.notebooks.feasibility import feasibility

The output.xlsx file contains a table of the molecules compared and their Tanimoto index.
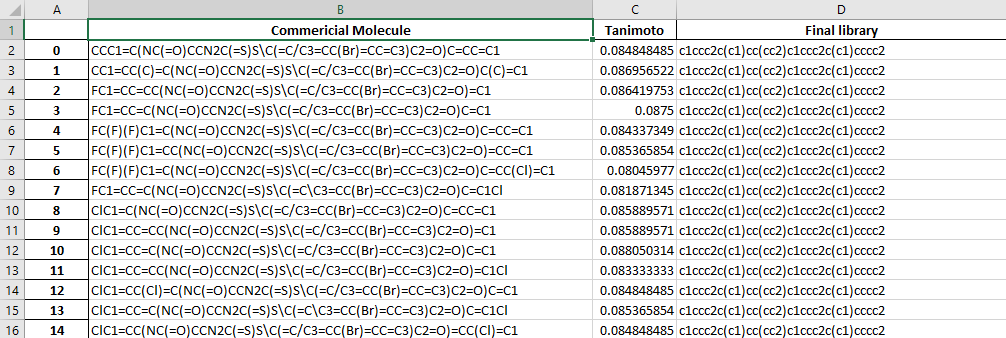
The dataframe.xlsx contains the table of molecule in the final library and the number of molecules which have a tanimoto index higher than the cut-off
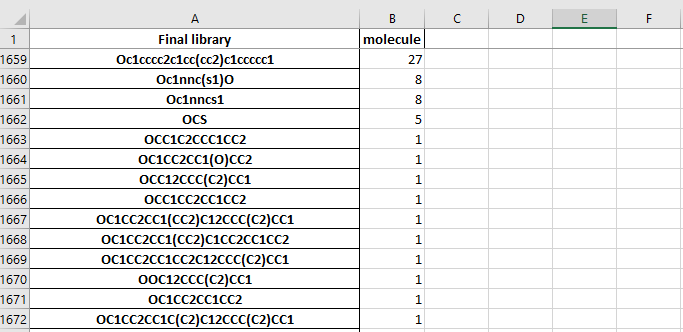
The stastictics.xlsx file cotains a table of the molecules and the number of each building block used in the molecule.
