Build a simple ML model
This page provides an example of a simple machine learning model developed using the GUI. We use the sample dataset from our library - this dataset has the SMILES codes for 500 compounds and their respective Highest Occupied Molecular Orbital (HOMO) energy in eV. The compounds are represented as Morgan fingerprints which are available through in the RDkit library. A 0.25 to 0.75 test-train split is used to split the data into the training data and the testing data.
The model is fitted to Morgan fingerprints and the HOMO energies of the training data. We fit the training data to a Multi-Layer Perceptron Regressor which is available through scikit-learn. We aim to predict the HOMO energies for the testing dataset.
[1]:
from chemml.wrapper.notebook import ChemMLNotebook
ui = ChemMLNotebook()
The computation graph will be displayed here:
The ChemML Wrapper's config file has been successfully saved ...
config file name: simple_ml_model.txt
current directory: c:\Users\nitin\Documents\UB\Hachmann_Group\chemml_dev_nitin\chemml\docs\ipython_notebooks
what's next? run the ChemML Wrapper using the config file with the following codes:
>>> from chemml.wrapper.engine import run
>>> run(INPUT_FILE = 'path_to_the_config_file', OUTPUT_DIRECTORY = 'CMLWrapper_out')
... you can also create a python script of the above codes and run it on any cluster that ChemML is installed.
The workflow gives a precise representation of all the intermediate steps, blocks used to develop the model, the saved data and the inputs/outputs to each task. Once the workflow is finalized, we save the input script with our desired file name in .txt format.
The GUI provides the file details and the steps to be followed to run the script - as shown below
Note: In this case, we specify our desired output directory as ‘Simple_ML_workflow’.
[3]:
from chemml.wrapper.engine import run
run(INPUT_FILE = './template_workflows/simple_ML_model.txt', OUTPUT_DIRECTORY = 'Simple_ML_workflow')
=================================================
=================================================
Tue Nov 5 14:58:38 2024
parsing the input file: ./template_workflows/simple_ML_model.txt ...
1 Task: (Input,datasets)
<<<<<<<
host = chemml
function = load_cep_homo
>>>>>>>
smiles -> send (id=0)
homo -> send (id=4)
:nothing to receive:
2 Task: (Output,file)
<<<<<<<
host = chemml
function = SaveFile
format = smi
header = False
filename = smiles
>>>>>>>
filepath -> send (id=1)
df <- recv (id=0)
3 Task: (Represent,molecular descriptors)
<<<<<<<
host = chemml
function = RDKitFingerprint
>>>>>>>
df -> send (id=2)
df -> send (id=3)
molfile <- recv (id=1)
4 Task: (Output,file)
<<<<<<<
host = chemml
function = SaveFile
filename = fps_rdkfp
>>>>>>>
:nothing to send:
df <- recv (id=2)
5 Task: (Prepare,split)
<<<<<<<
host = sklearn
function = train_test_split
>>>>>>>
dfx_train -> send (id=5)
dfy_train -> send (id=6)
dfx_test -> send (id=8)
dfy_test -> send (id=11)
dfx <- recv (id=3)
dfy <- recv (id=4)
6 Task: (Model,regression)
<<<<<<<
host = sklearn
function = MLPRegressor
func_method = fit
>>>>>>>
api -> send (id=7)
dfx <- recv (id=5)
dfy <- recv (id=6)
7 Task: (Model,regression)
<<<<<<<
host = sklearn
function = MLPRegressor
func_method = predict
>>>>>>>
dfy_predict -> send (id=9)
dfy_predict -> send (id=10)
api <- recv (id=7)
dfx <- recv (id=8)
8 Task: (Output,file)
<<<<<<<
host = chemml
function = SaveFile
filename = dfy_predict
>>>>>>>
:nothing to send:
df <- recv (id=9)
9 Task: (Visualize,plot)
<<<<<<<
host = chemml
function = scatter2D
x = 0
y = 0
>>>>>>>
fig -> send (id=13)
dfx <- recv (id=10)
dfy <- recv (id=11)
10 Task: (Output,figure)
<<<<<<<
host = chemml
function = SavePlot
filename = dfy_actual_vs_dfy_predict
output_directory = .
>>>>>>>
:nothing to send:
fig <- recv (id=12)
11 Task: (Visualize,artist)
<<<<<<<
host = chemml
function = decorator
title = true vs. predicted HOMO energy
xlabel = predicted HOMO energy (eV)
ylabel = true HOMO energy (eV)
grid = True
grid_color = g
size = 18
>>>>>>>
fig -> send (id=12)
fig <- recv (id=13)
=================================================
======= block#1: (chemml, load_cep_homo)
| run ...
| ... done!
| execution time: 5.36s (0h 0m 5.36s)
=======
======= block#2: (chemml, SaveFile)
| run ...
| ... done!
| execution time: 0.01s (0h 0m 0.01s)
=======
======= block#3: (chemml, RDKitFingerprint)
| run ...
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
[14:58:44] DEPRECATION WARNING: please use MorganGenerator
| ... done!
| execution time: 0.54s (0h 0m 0.54s)
=======
======= block#4: (chemml, SaveFile)
| run ...
| ... done!
| execution time: 0.09s (0h 0m 0.09s)
=======
======= block#5: (sklearn, train_test_split)
| run ...
| ... done!
| execution time: 0.08s (0h 0m 0.08s)
=======
======= block#6: (sklearn, MLPRegressor)
| run ...
c:\Users\nitin\anaconda3\envs\chemml_dev_env\Lib\site-packages\sklearn\neural_network\_multilayer_perceptron.py:1631: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().
y = column_or_1d(y, warn=True)
| ... done!
| execution time: 2.17s (0h 0m 2.17s)
=======
======= block#7: (sklearn, MLPRegressor)
| run ...
| ... done!
| execution time: 0.01s (0h 0m 0.01s)
=======
======= block#8: (chemml, SaveFile)
| run ...
| ... done!
| execution time: 0.00s (0h 0m 0.00s)
=======
======= block#9: (chemml, scatter2D)
| run ...
| ... done!
| execution time: 0.02s (0h 0m 0.02s)
=======
======= block#11: (chemml, decorator)
| run ...
| ... done!
| execution time: 0.00s (0h 0m 0.00s)
=======
======= block#10: (chemml, SavePlot)
| run ...
The Plot has been saved at: Simple_ML_workflow\./dfy_actual_vs_dfy_predict.png
| ... done!
| execution time: 0.09s (0h 0m 0.09s)
=======
Total execution time: 8.38s (0h 0m 8.38s)
2024-11-05 14:58:47
We create a plot to compare the predicted values of the HOMO enegeries to the actual values of the HOMO energies.
[4]:
from IPython.display import Image
Image(filename='Simple_ML_workflow/./dfy_actual_vs_dfy_predict.png')
[4]:
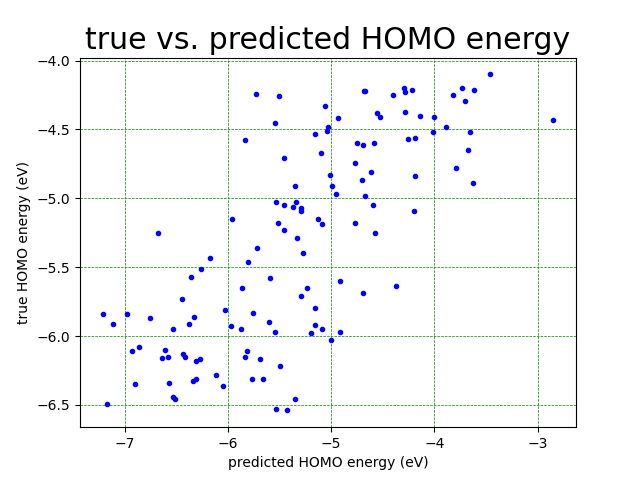
We plot a parity plot to comapre the model’s predicted values vs. the actual values. If the predicted results were fully accurate, we would have obtained clustering of the points along the equation y=x.